| |
|
| Products > VLifeMDS > LeadGrow |
|
|
|
|
|
|
| LeadGrow provides features for focused combinatorial library generation and screening to grow a lead molecule and generate large number of possibilities for lead optimization. Strong capabilities in generating combinatorial libraries and an extended Lipinski screen make LeadGrow a complete package for lead discovery and optimization. Seamless interfacing with VLifeQSAR enables the user to directly predict the activity of the library of molecules while a kNN MFA based QSAR model can be used to optimize and screen the library thus completing the full start to end capability of VLifeMDS in the ligand based design approach. |
| |
 |
| |
|
Focused library generation using descriptor screening
LeadGrow enables design of focused libraries by combining a strong library generation algorithm with an extended Lipinski screen. The library generation feature provides users with a significantly large potential chemical space for exploration due to LeadGrow’s ability to define multiple sites for substitution as well as the ability to define sites for bidentate substitutions. An extended Lipinski’s screen plus the facility to rank each molecule based on the score on the screen results in a library that is focused and optimized. |
Activity prediction of generated library
Seamless interfacing with VLifeQSAR allows for a direct prediction of activity of the generated molecules by using the QSAR model generated earlier on a similar set of molecules. This not only helps to design new molecules but also serves as an efficacy based screening of the generated molecules. |
Diversity analysis
Diversity of the generated molecules can be easily analyzed from the easy to use worksheet framework of LeadGrow. A choice of graphing options help to analyze the diversity. |
|
|
|
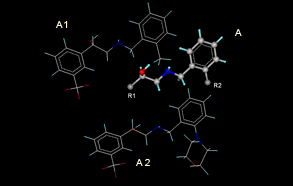 |
| Enumaration of combinotorial library using template based method (A-Template, A1& A2-Products) |
|
|
|
Peer reviewed algorithms
Algorithms in VLifeMDS have wide acceptance in scientific community. These algorithms are either published or are patent pending. There are multiple case studies of research conducted with VLifeMDS. |
Ease of plug-in / plug-out
Although an integrated platform, VLifeMDS has a modular architecture. Customers can buy the whole suite in one-go or buy incrementally as per the evolving needs of their research projects. Incremental module additions seamlessly integrate with rest of the modules to provide a consistent experience. |
Ease of customization & integration
VLifeMDS is developed fully in-house with complete ownership of every line of code. This provides unprecedented flexibility to add-on/ customize the product to suit customer specifications as well as integrating it to customers' existing discovery workbench. |
|
|
|
|
| |
|
 |
Pharmacophore identification and lead optimization for novel antifungals View |
 |
Anti-cancer : AKT1 QSAR model development View |
 |
Anti-cancer : Classification model development View |
|
|
|
|
|
|
|
|
|
|
|
|
|
| Conformer, force field analysis |
|
|
|
| |
|
|
|
| Combinatorial library generation |
|
|
|
|
|
|
|
|