| |
|
| Products > VLifeMDS > MolSign |
|
|
|
|
|
|
MolSign is a complete module for pharmacophore identification and modeling. Identification of the pharmacophoric features such as
H-bond donor, H-bond acceptor, positive charge, negative charge and hydrophobe facilitate the complete pharmacophoric mapping of the molecule, which can be further used for querying databases as a pharmacophore based search. MolSign easily interfaces with other modules of VLifeMDS such as ChemDBS, VLifeQSAR, ChemDBS and LeadGrow to complete a ligand based discovery package. |
| |
 |
| |
|
Feature extraction
The advanced Feature extraction function within MolSign considers hydrogen bond donor, hydrogen bond acceptor, negative charge group, positive charge group, aromatic ring center and aliphatic hydrophobe center as features to be extracted and used for pharmacophore identification thus leading to a comprehensive pharmacophoric model generation for the molecule. |
Pattern identification
Users can match the extracted features from different ligand molecules and use them for pattern search for a 3 point, 4 point, 5 point and up to n point pharmacophore identification on their own library of molecule. |
Strong visual display
MolSign utilizes the graphical display functions of VLife Engine to render high quality images of the selected pharmacophore and distribution of the pharmacophoric features around a set of molecules along with alignment of molecules based on selected features. |
|
|
|
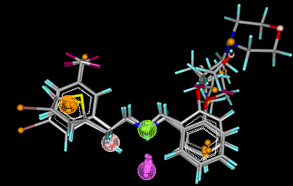 |
| Common feature pharmcophore mapping of antibacterial compounds |
|
|
|
Peer reviewed algorithms
Algorithms in VLifeMDS have wide acceptance in scientific community. These algorithms are either published or are patent pending. There are multiple case studies of research conducted with VLifeMDS. |
Ease of plug-in / plug-out
Although an integrated platform, VLifeMDS has a modular architecture. Customers can buy the whole suite in one-go or buy incrementally as per the evolving needs of their research projects. Incremental module additions seamlessly integrate with rest of the modules to provide a consistent experience. |
Ease of customization & integration
VLifeMDS is developed fully in-house with complete ownership of every line of code. This provides unprecedented flexibility to add-on/ customize the product to suit customer specifications as well as integrating it to customers' existing discovery workbench. |
|
|
|
|
| |
|
 |
Development and screening of NMDA agonist View |
 |
Application of enzymes to enhance drug action View |
 |
Pharmacophore identification and lead optimization for novel antifungals View |
|
|
|
|
|
|
|
|
|
|
|
|
|
| Conformer, force field analysis |
|
|
|
| |
|
|
| LeadGrow |
| Combinatorial library generation |
|
|
|
|
|
|
|
|