| |
| |
| |
|
|
| |
Physicochemical descriptors are based on the physicochemical properties of molecule. Sub classes of
hysicochemical descriptors are as follows.
1. Individual
2. Retention Index (chi)
3. Atomic valence connectivity index (chiv)
4. Path Count
5. Chi Chain
6. Chiv Chain
7. Chain Path Count
8. Cluster
9. Path Cluster
10. Kappa
11. Element Count
12. Dipole Moment
13. Electrostatic
14. Distance Based Topological
15. Estate numbers
16. Estate Contributions
17. Information Theory Index
18. Semi Empirical
19. Hydrophobicity XlogpA
20. Hydrophobicity XlogpK
21. Hydrophobicity SlogpA
22. Hydrophobicity SlogpK
23. Polar Surface Area |
|
| Various physicochemical descriptors are as follows. |
- Sub class: Individual
i. Mol.Wt: This descriptor signifies molecular weight of a compound.
ii. Volume: This descriptor signifies volume of a compound.
iii. H-AcceptorCount: Number of hydrogen bond acceptor atoms
iv. H-DonorCount: : Number of hydrogen bond donor atoms
v. RotatableBondCount: Number of rotatable bonds
vi. XlogP: This descriptor signifies ratio of solute concentration in octanol & water and generally
termed as Octanol Water partition Coefficient. This is atom based evaluation of logP as described
in Wang et al.)
vii. slogp: This descriptor signifies log of the octanol/water partition coefficient (including implicit
hydrogens). This property is an atomic contribution model [Crippen 1999] that calculates logP
from the given structure;i.e., the correct protonation state
viii. smr: This decriptor evaluates molecular refractivity (including implicit hydrogens) which also
measure of molecular size. This property is an atomic contribution model [Crippen 1999] that
assumes the correct protonation state (washed structures).
ix. polarizabilityAHC: This descriptor evaluates molecular polarizability using sum of atomic
polarizabilities using the atomic hybrid component (AHC).
x. polarizabilityAHP: This descriptor evaluates molecular polarizability using atomic hybrid
polarizability (AHP).
- Sub class: Chi
i. chi0: This descriptor signifies a retention index (zero order)derived directly from gradient
retention times
ii. chi1: This descriptor signifies a retention index (first order)derived directly from gradient
retention times
iii. chi2: This descriptor signifies a retention index (second order) derived directly from gradient
retention times.
iv. chi3: This descriptor signifies a retention index (third order)derived directly from gradient
retention times
v. chi4: This descriptor signifies a retention index (fourth order) derived directly from gradient
retention times.
vi. chi5: This descriptor signifies a retention index (fifth order) derived directly from gradient
retention times.
- Sub class: Chiv
i. chiV0: This descriptor signifies atomic valence connectivity index (order 0) from [Hall 1991]
and [Hall 1997]. This is calculated as the sum of 1/sqrt(vi) over all heavy atoms i with vi > 0.
ii. chiV1: This descriptor signifies atomic valence connectivity index (order 1) from [Hall 1991]
and [Hall 1997]. This is calculated as the sum of 1/sqrt(vivj) over all bonds between heavy atoms
i and j where i < j.
iii. chiV2: This descriptor signifies atomic valence connectivity index (order 2) from [Hall 1991]
and [Hall 1997].
iv. chiV3: This descriptor signifies atomic valence connectivity index (order 3) from [Hall 1991]
and [Hall 1997].
v. chiV4: This descriptor signifies atomic valence connectivity index (order 4) from [Hall 1991]
and [Hall 1997].
vi. chiV5: This descriptor signifies atomic valence connectivity index (order 5) from [Hall 1991]
and [Hall 1997].
- Sub class: Path Count
i. 0PathCount: This descriptor signifies total number of fragments of zero order (atoms) in a
compound.
ii. 1PathCount: This descriptor signifies total number of fragments of first order (bonds) in a
compound.
iii. 2PathCount: This descriptor signifies total number of fragments of second order (two bond path)
in a compound.
iv. 3PathCount: This descriptor signifies total number of fragments of third order (three bond path)
in a compound.
v. 4PathCount: This descriptor signifies total number of fragments of fourth order (four bond path)
in a compound.
vi. 5PathCount: This
- Sub class: Chi Chain
i. chi3chain: This descriptor signifies a retention index for three membered ring.
ii. chi4chain: This descriptor signifies a retention index for four membered ring.
iii. chi5chain: This descriptor signifies a retention index for five membered ring.
iv. chi6chain: This descriptor signifies a retention index for six membered ring.
- Sub class: Chiv Chain
i. chiV3chain: This descriptor signifies atomic valence connectivity index for three membered ring.
ii. chiV4chain: This descriptor signifies atomic valence connectivity index for four membered ring.
iii. chiV5chain: This descriptor signifies atomic valence connectivity index for five membered ring
iv. chiV6chain: This descriptor signifies atomic valence connectivity index for six membered ring
- Sub class: Chain Path count
i. 3ChainCount: This descriptor signifies total number three membered rings in a compound.
ii. 4ChainCount: This descriptor signifies total number four membered rings in a compound.
iii. 5ChainCount: This descriptor signifies total number five membered rings in a compound.
iv. 6ChainCount: This descriptor signifies total number six membered rings in a compound.
- Sub class: Cluster
i. chi3Cluster: This descriptor signifies simple 3rd order cluster chi index in a compound.
ii. chiV3Cluster: This descriptor signifies valence molecular connectivity index of 3rd order cluster.
iii. 3ClusterCount: This descriptor signifies total number of fragments of third order cluster in a
molecule.
- Sub Class: Path Cluster
i. chi4pathCluster: This descriptor signifies molecular connectivity index of 4th order pathcluster.
ii. chiV4pathCluster: This descriptor signifies valence molecular connectivity index of 3rd order
pathcluster.
iii. 4pathClusterCount: This descriptor signifies total number of fragments of fourth order
pathcluster in a molecule.
- Sub Class: Kappa
i. kappa1: This descriptor signifies first kappa shape index: (n-1)2 / m2 [Hall 1991]
ii. kappa2: This descriptor signifies second kappa shape index: (n-1)2 / m2 [Hall 1991]
iii. kappa3: This descriptor signifies third kappa shape index: (n-1) (n-3)2 / p32 for odd n, and (n-
3) (n-2)2 /p32 for even n [Hall 1991]
iv. k1alpha: This descriptor signifies first alpha modified shape index: s (s-1)2 / m2 where s = n + a
[Hall 1991]
v. k2alpha: This descriptor signifies second alpha modified shape index: s (s-1)2 / m2 where
s = n + a [Hall 1991]
vi. k3alpha: This descriptor signifies third alpha modified shape index: (n-1) (n-3)2 / p32 for odd n,
and (n-3) (n-2)2 / p32 for even n where s = n + a [Hall 1991]
- Sub Class: Element Count
i. HydrogensCount: This descriptor signifies number of hydrogen atoms in a compound.
ii. CarbonsCount: This descriptor signifies number of carbon atoms in a compound.
iii. SulfursCount : This descriptor signifies number of sulphur atoms in a compound.
iv. OxygensCount: This descriptor signifies number of oxygen atoms in a compound.
v. NitrogensCount: This descriptor signifies number of nitrogen atoms in a compound.
vi. ChlorinesCount: This descriptor signifies number of chlorine atoms in a compound.
vii. FluorinesCount: This descriptor signifies number of fluorine atoms in a compound.
viii. BrominesCount: This descriptor signifies number of bromine atoms in a compound.
ix. IodinesCount: This descriptor signifies number of iodine atoms in a compound.
- Sub Class: Dipole Moment
i. XcompDipole : This descriptor signifies the x component of the dipole moment (external
coordinates).
ii. YcompDipole : This descriptor signifies the y component of the dipole moment (external
coordinates).
iii. ZcompDipole : This descriptor signifies the z component of the dipole moment (external
coordinates).
iv. DipoleMoment: This descriptor signifies dipole moment calculated from the partial charges of
the molecule.
v. Quadrupole1: This descriptor signifies magnitude of first tensor of quadrupole moments.
vi. Quadrupole2: This descriptor signifies magnitude of second tensor of quadrupole moments.
vii. Quadrupole3: This descriptor signifies magnitude of third tensor of quadrupole moments.
- Sub class: Electrostatic
i. vdWSurfaceArea: This descriptor signifies total van der Waals surface area of the molecule.
ii. +vePotentialSurfaceArea: This descriptor signifies total van der Waals surface area with positive
electrostatic potential of the molecule.
iii. -vePotentialSurfaceArea: This descriptor signifies total van der Waals surface area with negative
electrostatic potential of the molecule.
iv. Most+vePotential: This descriptor signifies the highest value of +ve electrostatic potential on van
der Waals surface area of the molecule.
v. Most-vePotential: This descriptor signifies the highest value of -ve electrostatic potential on van
der Waals surface area of the molecule.
vi. AveragePotential: This descriptor signifies average of the total electrostatic potential on van der
Waals surface area of the molecule.
vii. Average+vePotential: This descriptor signifies the average of the total +ve electrostatic potential
on van der Waals surface area of the molecule.
viii. Average-vePotential: This descriptor signifies the average of the total -ve electrostatic potential
on van der Waals surface area of the molecule.
ix. Most+ve&-vePotentailDistance: This descriptor signifies the distance between points having the
highest value of +ve and highest value of –ve electrostatic potential on van der Waals surface
area of the molecule.
- Sub Class: Distance based Topological
i. DistTopo: This descriptor signifies distance based topological index.
ii. ConnectivityIndex: This signifies a numeric descriptor derived from molecular topology.
iii. WienerIndex : This descriptor signifies the sum of the numbers of edges in shortest paths in a
chemical graph between all pairs of non-hydrogen atoms in a molecule.
iv. RadiusOfGyration: This descriptor signifies size descriptor for the distribution of atomic masses
in a molecule.
v. MomInertiaX : This descriptor signifies moment of interia at X-axix
vi. MomInertiaY : This descriptor signifies moment of interia at Y-axix
vii. MomInertiaZ : This descriptor signifies moment of interia at Z-axix
viii. BalabanIndexJ:
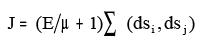
Where dsi, dsj = sum of the row i and j of the distance matrix
E = number of edges
µ = Number of rings in a molecule.
ix. BalabanB:
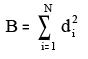
Where di = Number of vertices deleted at each step
N = Number of all vertices
x. BalabanC:
C = (1/ 2)(B - 2N + U)
Where B = Balaban B index
N = Number of all vertices
U = [1-(-1)N]
xi. BalabanQ:
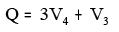
Where V3 = Number of vertices of degree 3
V4 = Number of vertices of degree 4
xii. BalabanCdash:
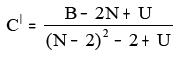
Where B = Balaban B index
N = Number of all vertices and U = [1-(-1)N]
xiii. BalabanQdash:
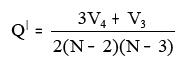
Where V3 = Number of vertices of degree 3
V4 = Number of vertices of degree 4
N = Number of all vertices
xiv. HosoyaIndex: This descriptor signifies the topological index or Z index of a graph is the total
number of matching in it plus 1 ("plus 1" accounts for the number of matchings with 0 edges).
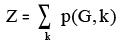
Where, p(G,k) = Number of ways in which K edges from all bonds of a graph G may be chosen
so that no two of them are adjacent
- Sub class: Estate Numbers
i. SsCH3count: This descriptor defines the total number of –CH3 group connected with single bond
ii. SdCH2count: This descriptor defines the total number of –CH2 group connected with double
bond
iii. SssCH2count : This descriptor defines the total number of –CH2 group connected with two
single bonds
iv. StCHcount: This descriptor defines the total number of –CH group connected with triple bond
v. SdsCHcount: This descriptor defines the total number of –CH group connected with one double
and one single bond.
vi. SaaCHcount: This descriptor defines the total number of carbon atoms connected with a
hydrogen along with two aromatic bonds.
vii. SsssCHcount: This descriptor defines the total number of –CH group connected with three single
bond.
viii. SddCcount: This descriptor defines total number of carbon atoms (= C =) with two double bonds
present in the molecule.
ix. StsCcount: This descriptors defines total number of carbon atoms (- C  ) with a triple bond and
a single bond present in the molecule. ) with a triple bond and
a single bond present in the molecule.
x. SdssCcount: This descriptor defines the total number of carbon connected with one double and
two single bond.
xi. SaasCcount: This descriptor defines the total number of carbon connected with one single bond
along with two aromatic bonds.
xii. SaaaCcount: This descriptor defines the total number of carbon connected with three aromatic
bonds.
xiii. SssssCcount: This descriptor defines the total number of carbon connected with four single
bonds.
xiv. SsNH3count: This descriptor defines the total number of –NH3 group connected with one
single bond.
xv. SsNH2count: This descriptor defines the total number of –NH2 group connected with one
single bond.
xvi. SssNH2count : This descriptor defines the total number of –NH2 group connected with two
single bonds.
xvii. SdNHcount: This descriptor defines the total number of –NH group connected with one double
bond.
xviii. SssNHcount: This descriptor defines the total number of –NH group connected with two single
bond.
xix. SaaNHcount: This descriptor defines the total number of –NH group connected with two
aromatic bonds.
xx. StNcount: This descriptor defines the total number of nitrogen connected with triple bond.
xxi. SsssNHcount: This descriptor defines the total number of –NH group connected with three
single bonds.
xxii. SdsNcount: This descriptor defines the total number of nitrogen connected with one single and
one double bond.
xxiii. SaaNcount: This descriptor defines the total number of nitrogen connected with two aromatic
bonds.
xxiv. SsssNcount: This descriptor defines the total number of nitrogen connected with three single
bonds.
xxv. SddsN(nitro)count: This descriptor defines the total number of nitro group connected with one
single and two double bonds.
xxvi. SaasN(Noxide)count : This descriptor defines the total number of nitro oxide group connected
with one single along with two aromatic bonds.
xxvii. SssssN(onium)count : This descriptor defines the total number of N- onium group connected
with four single bonds.
xxviii. SsOHcount: This descriptor defines the total number of –OH group connected with one single
bond.
xxix. SdOcount: This descriptor defines the total number of oxygen connected with one double
bond.
xxx. SssOcount: This descriptor defines the total number of oxygen connected with two single
bonds.
xxxi. SaaOcount: This descriptor defines the total number of oxygen connected with two aromatic
bonds.
xxxii. SsPH2count: This descriptor defines the total number of –PH2 group connected with one
single bond.
xxxiii. SssPHcount: This descriptor defines the total number of –PH group connected with two
single bonds.
xxxiv. SsssPcount: This descriptor defines the total number of phosphorous atom connected with
three single bonds.
xxxv. SdsssPcount: This descriptor defines the total number of phosphorous atom connected with
three single bonds and one double bond.
xxxvi. SsssssPcount: This descriptor defines the total number of phosphorous atom connected with
five single bonds.
xxxvii. SsSHcount: This descriptor defines the total number of –SH group connected with one single
bond.
xxxviii. SdScount: This descriptor defines the total number of sulphur atom connected with one
double bond.
xxxix. SssScount: This descriptor defines the total number of sulphur atom connected with two
single bonds.
xl. SaaScount: This descriptor defines the total number of sulphur atom connected with two
aromatic bonds.
xli. SdssS(sulfone)count : This descriptor defines the total number of sulphone group connected
with two single and one double bond.
xlii. SddssS(sulfate)count: This descriptor defines the total number of sulphate group connected with
two single and two double bonds.
xliii. SsClcount: This descriptor defines the total number of chlorine atom connected with one single
bond.
xliv. SsBrcount: This descriptor defines the total number of bromine atom connected with one single
bond.
xlv. SsIcount: This descriptor defines the total number of iodine atom connected with one single
bond.
xlvi. SsFcount: This descriptor defines the total number of fluorine atom connected with one single
bond.
- Sub class: Estate contributions
i. SsCH3E-index: Electrotopological state indices for number of -CH3 group connected with one
single bond.
ii. SdCH2E-index: Electrotopological state indices for number of –CH2 group connected with one
double bond.
iii. SssCH2E-index: Electrotopological state indices for number of –CH2 group connected with two
single bonds.
iv. StCHE-index: Electrotopological state indices for number of –CH group connected with one
triple bond
v. SdsCHE-index: Electrotopological state indices for number of –CH group connected with one
double and one single bond.
vi. SaaCHE-index: Electrotopological state indices for number of –CH group connected with two
aromatic bonds.
vii. SsssCHE-index: Electrotopological state indices for number of –CH group connected with three
single bonds.
viii. SddCE-index: Electrotopological state indices for number of carbon atom connected with two
double bonds.
ix. StsCE-index: Electrotopological state indices for number of carbon atom connected with one
triple and one single bond.
x. SdssCE-index : Electrotopological state indices for number of carbon atom connected with
one double and two single bonds.
xi. SaasCE-index : Electrotopological state indices for number of carbon atom connected with
one single bond along with two aromatic bonds.
xii. SaaaCE-index : Electrotopological state indices for number of carbon atom connected with
three aromatic bonds.
xiii. SssssCE-index: Electrotopological state indices for number of carbon atom connected with four
single bonds.
xiv. SsNH3E-index: Electrotopological state indices for number of –NH3 group connected with one
single bond.
xv. SsNH2E-index: Electrotopological state indices for number of –NH2 group connected with one
single bond.
xvi. SssNH2E-index: Electrotopological state indices for number of –NH2 group connected with two
single bond.
xvii. SdNHE-index : Electrotopological state indices for number of –NH group connected with one
double bond.
xviii. SssNHE-index : Electrotopological state indices for number of –NH group connected with two
single bonds.
xix. SaaNHE-index: Electrotopological state indices for number of –NH group connected with two
aromatic bonds.
xx. StNE-index: Electrotopological state indices for number of nitrogen atom connected with one
triple bonds.
xxi. SsssNHE-index: Electrotopological state indices for number of –NH group connected with three
single bonds.
xxii. SdsNEindex: Electrotopological state indices for number of nitrogen atom connected with two
double and one single bond.
xxiii. SaaNE-index: Electrotopological state indices for number of nitrogen atom connected with two
aromatic bonds.
xxiv. SsssNE-index: Electrotopological state indices for number of nitrogen atom connected with
three single bonds.
xxv. SddsN(nitro)E-index : Electrotopological state indices for number of –nitro group connected
with two double and one single bond.
xxvi. SaasN(Noxide)E-index: Electrotopological state indices for number of nitro-oxide group
connected with two aromatic and one single bond.
xxvii. SssssN(onium)E-index: Electrotopological state indices for number of N-onium group
connected with four single bonds.
xxviii. SsOHE-index: Electrotopological state indices for number of –OH group connected with one
single bond.
xxix. SdOE-index: Electrotopological state indices for number of oxygen atom connected with one
double bond.
xxx. SssOE-index: Electrotopological state indices for number of oxygen atom connected with two single
bonds.
xxxi. SaaOE-index: Electrotopological state indices for number of oxygen atom connected with two
aromatic bonds.
xxxii. SsPH2E-index : Electrotopological state indices for number of –PH2 group connected
with one single bond.
xxxiii. SssPHE-index : Electrotopological state indices for number of –PH group connected
with two single bonds.
xxxiv. SsssPE-index: Electrotopological state indices for number of phosphorous atom connected
with three single bonds.
xxxv. SdsssPE-index: Electrotopological state indices for number of phosphorous atom connected
with three single bonds along with one double bond.
xxxvi. SsssssPE-index: Electrotopological state indices for number of phosphorous atom connected
with five single bonds.
xxxvii. SsSHE-index: Electrotopological state indices for number of –SH group connected with one
single bond.
xxxviii. SdSE-index: Electrotopological state indices for number of sulphur atom connected with
one double bond.
xxxix. SssSE-index: Electrotopological state indices for number of sulphur atom connected with two
single bonds.
xl. SaaSE-index: Electrotopological state indices for number of sulphur atom connected with two
aromatic bonds.
xli. SdssS(sulfone)E-index: Electrotopological state indices for number of sulfone group connected
with two single bonds.
xlii. SddssS(sulfate)E-index: Electrotopological state indices for number of sulfate group connected
with two single bonds and two double bonds.
xliii. SsClE-index: Electrotopological state indices for number of chlorine connected with one single
bond.
xliv. SsBrE-index: Electrotopological state indices for number of bromine connected with one single
bond.
xlv. SsIE-index: Electrotopological state indices for number of iodine connected with one single
bond.
xlvi. SsFE-index: Electrotopological state indices for number of fluorine connected with one single
bond.
- Sub class: Information theory based
i. Ipc: This is a type of information theory based descriptors.
ii. IpcAverage: This is a type of information theory based descriptors.
iii. Id: This is a type of information theory based descriptors.
iv. IdAverage: This is a type of information theory based descriptors.
v. Idw: This is a type of information-based descriptors.
vi. IdwAverage: This is a type of information-based descriptors.
- Sub Class: Semi empirical
i. HUMOEnergy: This descriptor signifies energy of highest occupied molecular orbital.
ii. LUMOEnergy: This descriptor signifies energy of highest unoccupied molecular orbital.
iii. HeatOfFormation: This descriptor signifies the heat of formation of a compound.
iv. IonizationPotential: This descriptor signifies ionization potential of a compound.
v. SumOfAbsoluteCharges:
vi. QMDipoleX: Induced dipole moment along X-axis
vii. QMDipoleY: Induced dipole moment along Y-axis
viii. QMDipoleZ: Induced dipole moment along Z-axis
ix. QMDipoleMagnitude: Magnitude of induced dipole moment.
x. XXPolarizability: Induced polarizability along XX axis
xi. YYPolarizability: Induced polarizability along YY axis
xii. ZZPolarizability: Induced polarizability along ZZ axis
xiii. XYPolarizability: Induced polarizability along XY axis
xiv. XZPolarizability: Induced polarizability along XZ axis
xv. YZPolarizability: Induced polarizability along YZ axis
xvi. AveragePolarizability: Average induced polarizability along all axis.
- Sub class: Hydrophobicity XlogpA
i. XAHydrophobicArea: vdW surface descriptor showing hydrophobic surface area. (By Audry
Method using Xlogp)
ii. XAHydrophilicArea: vdW surface descriptor showing hydrophilic surface area. (By Audry
Method using Xlogp)
iii. XAMostHydrophobic: Most hydrophobic value on the vdW surface. (By Audry Method using
Xlogp)
iv. XAMostHydrophilic: Most hydrophilic value on the vdW surface. (By Audry Method using
Xlogp)
v. XAAverage: Average hydophobicity function value. (By Audry Method using Xlogp)
vi. XAAverageHydrophobicity: Average hydrophobic value on the vdW surface. (By Audry Method
using Xlogp)
vii. XAAverageHydrophilicity: Average hydrophilic value on the vdW surface. (By Audry Method
using Xlogp)
viii. XAMostHydrophobicHydrophilicDistance: This descriptor signifies distance between most
hydrophobic and hydrophilic point on the vdW surface. (By Audry Method using Xlogp)
- Sub class: Hydrophobicity XlogpK
i. XKHydrophobicArea: vdW surface descriptor showing hydrophobic surface area. (By Kellog
Method using Xlogp)
ii. XKHydrophilicArea: vdW surface descriptor showing hydrophilic surface area. (By Kellog
Method using Xlogp)
iii. XKMostHydrophobic: Most hydrophobic value on the vdW surface. (By Kellog Method using
Xlogp)
iv. XKMostHydrophilic: Most hydrophilic value on the vdW surface. (By Kellog Method using
Xlogp)
v. XKAverage: Average hydophobicity function value. (By Kellog Method using Xlogp)
vi. XKAverageHydrophobicity: Average hydrophobic value on the vdW surface. (By Kellog
Method using Xlogp)
vii. XKAverageHydrophilicity: Average hydrophilic value on the vdW surface. (By Kellog Method
using Xlogp)
viii. XKMostHydrophobicHydrophilicDistance: This descriptor signifies distance between most
hydrophobic and hydrophilic point on the vdW surface. (By Kellog Method using Xlogp)
- Sub class: Hydrophobicity SlogpA
i. SAHydrophobicArea: vdW surface descriptor showing hydrophobic surface area. (By Audry
Method using Slogp)
ii. SAHydrophilicArea: vdW surface descriptor showing hydrophilic surface area. (By Audry
Method using SlogP)
iii. SAMostHydrophobic: Most hydrophobic value on the vdW surface. (By Audry Method using
Slogp)
iv. SAMostHydrophilic: Most hydrophilic value on the vdW surface. (By Audry Method using
Slogp)
v. SAAverage: Average hydophobicity function value. (By Audry Method using Slogp)
vi. SAAverageHydrophobicity: Average hydrophobic value on the vdW surface. (By Audry Method
using Slogp)
vii. SAAverageHydrophilicity: Most hydrophilic value on the vdW surface. (By Audry Method using
Slogp)
viii. SAMostHydrophobicHydrophilicDistance: This descriptor signifies distance between most
hydrophobic and hydrophilic point on the vdW surface. (By Audry Method using Slogp)
- Sub class: Hydrophobicity SlogpK
i. SKHydrophobicArea: vdW surface descriptor showing hydrophobic surface area. (By Kellog
Method using Slogp)
ii. SKHydrophilicArea: vdW surface descriptor showing hydrophilic surface area. (By Kellog
Method using Slogp)
iii. SKMostHydrophobic: Most hydrophobic value on the vdW surface. (By Kellog Method using
Slogp)
iv. SKMostHydrophilic: Most hydrophilic value on the vdW surface. (By Kellog Method using
Slogp)
v. SKAverage: Average hydophobicity function value. (By Kellog Method using Slogp)
vi. SKAverageHydrophobicity: Average hydrophobic value on the vdW surface. (By Kellog Method
using Slogp)
vii. SKAverageHydrophilicity: Average hydrophilic value on the vdW surface. (By Kellog Method
using Slogp)
viii. SKMostHydrophobicHydrophilicDistance: This descriptor signifies distance between most
hydrophobic and hydrophilic point on the vdW surface. (By Kellog Method using Slogp)
- Sub class: Polar Surface Area
i. PolarSurfaceAreaExcludingPandS: This descriptor signifies total polar surface area excluding
phosphorous and sulphur.
ii. PolarSurfaceAreaIncludingPandS: This descriptor signifies total polar surface area including
phosphorous and sulphur.
|
|
| B. Alignment Independent (AI) descriptors class: (more than 700 descriptors) |
Alignment Independent descriptors are calculated as discussed in Baumann’s paper [1]. For calculation of
AI descriptors every atom in the molecule was assigned at least one and at most three attributes. The first
attribute is ‘T-attribute’ to thoroughly characterize the topology of the molecule. The second attribute is
the atom type. The atom symbol is used here. The third attribute is assigned to atoms taking part in a
double or triple bond. After all atoms have been assigned their respective attributes, selective distancecount statistics for all combinations of different attributes are computed [1]. A selective distance count
statistic ‘XY2’ (e.g. ‘TOPO2N3) counts all the fragments between start atom with attribute ‘X’ (e.g. ‘2’
double bonded atom) and end atom with attribute ‘Y’ (e.g. ‘N’) separated by the graph distance 3. The
graph distance can be defined as the smallest number of atoms along the path connecting two atoms in
molecular structure. In this study to calculate AI descriptors, we have used following attributes: 2 (double
bonded atom), 3(triple bonded atom), C, N, O, S, H, F, Cl, Br and I and the distance range of 0 to 7.
Some other examples are as follows:
i. T_2_O_7: This is the count of number of double bounded atoms (i.e. any double bonded atom,
T_2) separated from Oxygen atom by 7 bonds in a molecule.
ii. T_2_N_5: This is the count of number of double bounded atoms (i.e. any double bonded atom,
T_2) separated from Nitrogen atom by 5 bonds.
iii. T_N_N_5: This is the count of number of Nitrogen atoms (single double or triple bonded)
separated from any other Nitrogen atom (single double or triple bonded) by 5 bonds in a
molecule.
iv. T_2_2_6: This is the count of number of double bounded atoms (i.e. any double bonded atom,
T_2) separated from any other double bonded atom by 6 bonds in a molecule.
v. T_C_O_1: This is the count of number of Carbon atoms (single double or triple bonded)
separated from any Oxygen atom (single or double bonded) by 1 bond distance in a molecule.
vi. T_O_Cl_5: This is the count of number of Oxygen atoms (single double or triple bonded)
separated from Chlorine atom by 5 bond distance in a molecule.
Similarly around 700 alignment independent descriptors can be generated considering topology of the
molecule, atom type & bond. |
|
| C. Atom type count descriptors class: (total 99 descriptors) |
The atom type count descriptors are based on MMFF atom types and their count in each molecule. In
MMFF, there are 99 atom types and hence 99 descriptors indicating number of times that atom type has
occurred in a given molecule are generated.
Reference:
[1] A.T. Balaban, Chem. Phys. Letters, 89, 399-404, (1982).
[2] D. Plavsic, M. Soskic, N. Lers, J. Chem. Inf. Comput. Sci., 38, 889-892,(1998).
[3] H. Wiener, J. Am. Chem. Soc. 69, 17 (1947).
[4] M. Randic, in Encyclopedia of Computational Chemistry, P.v.R Schleyer, N. L. Allinger, T. Clark, J.
Gasteiger, P. A. Kollman, H. F. Schaefer III, P. R. Schreiner Eds, John Wiley & Sons, Chinchester,1998,
pp. 3018-3032
[5] D. Bonchev and Trinajstic, J. Chem. Phys., 67, 4517-4533, (1977).
[6] L.B. Kier and L.H. Hall, Eur. J. Med. Chem., 12, 307 (1977).
[7] L.H. Hall and L.B. Kier in Reviews of Computational Chemistry, Vol. 2, D.B. Boyd and K.
Lipkowitz, eds. (1991).
[8] L.B. Kier, Quant. Struct-Act. Relat., 4, 109 (1985).
[9] L.H. Hall, B.K. Mohney and L.B. Kier, J. Chem. Inf. Comput. Sci., 31, 76 (1991).
[10] L.H. Hall and L.B. Kier, J. Chem. Inf. Comput. Sci., 35, 1039-1045 (1995).
[11] R. Wang, Y. Fu, L. Lai, J. Chem. Inf. Comput. Sci., 37, 615-621 (1997).
[12] F.C. Wireko, G. E. Kellogg, D. J. Abraham, J. Med. Chem., 34, 758 (1991).
[13] E. Audry, J.P. Dubost, J.C. Colleter, P. Dallet, Eur. J. Med. Chem-Chim. Ther. 21, 71-72, (1986).
[14] S.A. Wildman, G.M. Crippen, J. Chem. Inf. Comput. Sci. 39, 868-873, (1999).
[15] K. Baumann, J. Chem. Inf. Comput. Sci., 42, 26-35 (2002). |
|
| |
| |
|
|
|
|
|
|