| |
|
| Technology > Aakar |
|
|
|
|
|
|
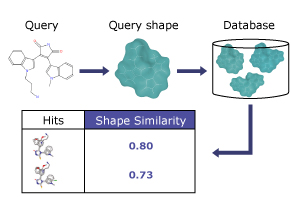
The ability to quickly and accurately identify similar molecular structures from a large database of molecules, equips a medicinal chemist with the right tool to realize a number of applications such as scaffold hopping, bioisostere replacement, virtual library design, and flexible ligand superimposition. Aakar provides a powerful and fast alignment independent shape search method with or without taking into consideration the chemical pharmacophoric features.
Features in Aakar
- Alignment independent shape descriptors
- Multiple shape descriptors:
- Topological arrangement of atoms
- Topological & Pharmacophoric feature based
- Characteristic vector based on spectral distance matrix
- Characteristic vector & Atomic charge based
|
| |
 |
| |
|
| GQSAR: A patent pending technology for fragment based QSAR developed by VLife which enhances use of QSAR for design optimization of molecule delivering highly specific site directed clues for design modification. |
| VLifeSCOPE: A novel technology application creating a hybrid approach for lead optimization and prioritization of design for a given purpose from a library of molecules. |
| kNN-MFA: A novel combination of k-nearest neighbor method with molecular field analysis which takes cognizance of critical non-linear relationships between molecular properties with its activity. |
| LeadGrow+: An extension to the combinatorial library generation capability of VLifeMDS that significantly expands the chemical universe by enabling template substitution. |
| VLifeAutoQSAR: Unique automated approach to conduct QSAR that provides a best result based on a consensus of multiple QSAR models generated. |
| VLifeWorkFlow: A tool to customize and automate the discovery protocols of users using the CADD components. |
|
|
 |
|
|
|
|
|
|
• Scaffold Hopping
• Bioisosteric Replacement
|
|
| |
|
| |
|
 |
Development and screening of NMDA agonist View |
 |
Target Identification for existing nutraceutical molecule View |
 |
Homology modeling using BioPredicta View |
|
|
|
|
|
|
|
|
"This new QSAR methodology gives QSARpro, a decisive edge over conventional QSAR. The ability to combine kNN with MFA is a unique approach which I came across only in QSARpro from VLife. It is now a method of choice in my research."
Dr. S.P.Gupta
Ex-BITS, Pilani |
|
|
| GQSAR |
| For site specific design clues |
|
|
|
| kNN-MFA |
| Taking cognizance of non-linearity |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| LeadGrow |
| Combinatorial library generation |
|
|
|
|
|
|
|
|
|