| |
|
| Technology > GQSAR |
|
|
|
|
|
|
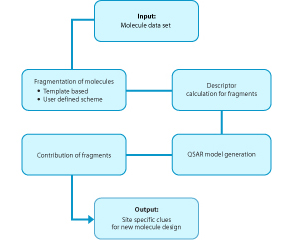
GQSAR is a breakthrough patent pending methodology that significantly enhances the use of QSAR as an approach for new molecule design. As a predictive tool for activity, this method is significantly superior to conventional 3D and 2D QSAR.
In this method, every molecule of the data set is considered as a set of fragments, the fragmentation scheme being either template based or user defined.
The descriptors are evaluated for each fragment and a relationship between these fragment descriptors is formed with the activity of the whole molecule.
Unlike conventional QSAR, with the GQSAR, researchers get critically important site specific clues within a molecule where a particular descriptor needs to be modified.
GQSAR approach builds upon the basic focus of QSAR by applying the knowledge gained in the field over the past four decades in terms of molecular descriptors, statistical modeling etc. |
| |
 |
| |
|
| VLifeSCOPE: A novel technology application creating a hybrid approach for lead optimization and prioritization of design for a given purpose from a library of molecules. |
| kNN-MFA: A novel combination of k-nearest neighbor method with molecular field analysis which takes cognizance of critical non-linear relationships between molecular properties with its activity. |
| LeadGrow+: An extension to the combinatorial library generation capability of VLifeMDS that significantly expands the chemical universe by enabling template substitution. |
| VLifeAutoQSAR: Unique automated approach to conduct QSAR that provides a best result based on a consensus of multiple QSAR models generated. |
| Aakar: A powerful and fast alignment independent shape search method with or without taking into consideration the chemical pharmacophoric features. |
| VLifeWorkFlow: A tool to customize and automate the discovery protocols of users using the CADD components. |
|
|
 |
| Enlarge |
|
Pub: QSAR & Combinatorial Science 2009
Group Based QSAR(GQSAR): Mitigating Interpretation Challenges in QSAR,
Subhash Ajmani, Kamalakar Jadhav, Sudhir A. Kulkarni |
|
|
|
Extends application of QSAR beyond just library screening
Provides very important site specific clues for better molecule design optimization |
Delivers understanding about the contribution of various parts of a molecule towards governing biological activity variation |
Offers the best of conventional QSAR approaches while mitigating interpretation challenges from such approaches |
Choice of fragmentation schemes based on the nature of the data set to handle congeneric as well as non-congeneric series of molecules |
|
|
|
| |
|
 |
A novel QSAR technology for new molecule design View |
 |
Development of dual active antifungals View |
 |
Anti-cancer : Classification model development View |
|
|
|
|
|
|
|
|
"This new QSAR methodology gives QSARpro, a decisive edge over conventional QSAR. The ability to combine kNN with MFA is a unique approach which I came across only in QSARpro from VLife. It is now a method of choice in my research."
Dr. S.P.Gupta
Ex-BITS, Pilani |
|
|
|
| For site specific design clues |
|
|
|
| kNN-MFA |
| Taking cognizance of non-linearity |
|
|
|
|
| Aakar |
| Shape Based Screening |
|
|
|
|
|
|
|
|
|
|
|
|
| LeadGrow |
| Combinatorial library generation |
|
|
|
|
|
|
|
|